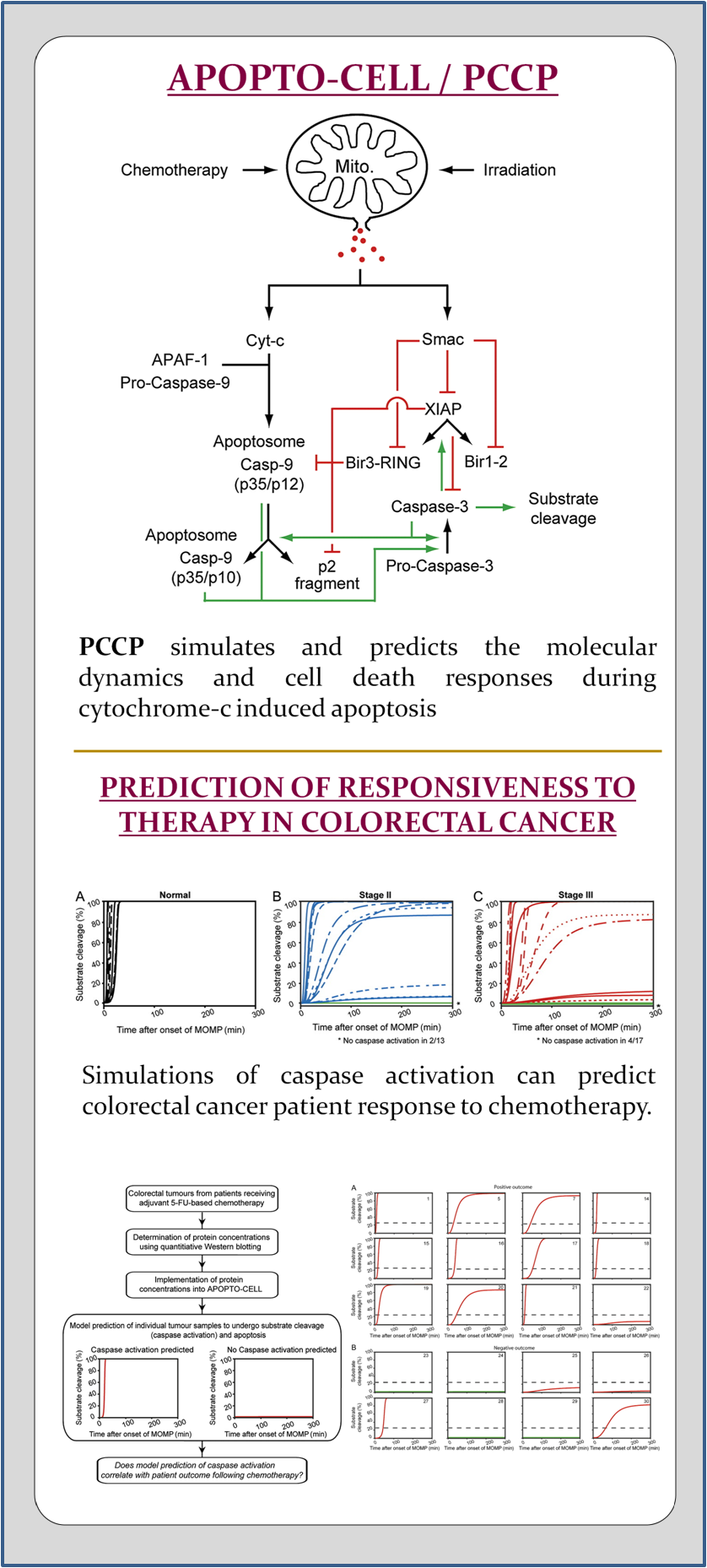
APOPTO-CELL is an mathematical systems model applied in a clinical workflow and software tool for the Prediction of Cancer response to genotoxic Chemotherapy and Personalised neoadjuvant treatments (PCCP).
APOPTO-CELL
Introduction
This model allows the rendering of the molecular dynamics and responses following cytochrome c-induced/apoptosome-dependent apoptosis. The molecular composition of the reaction network can either be manually defined or selected from quantitative data for specific cellular systems deposited in a database. To establish a versatile cellular model, users are invited to extend the database content by submitting data from own quantitative analyses.
Three different program modes can be selected:
Temporal Protein Profiles: This option allows the visualization of the temporal profiles of all proteins and protein complexes involved in the apoptotic process, including intermediary and transient products that cannot be monitored experimentally. Furthermore, the cleavage profile of intracellular effector caspase substrate is displayed and can directly be compared to experimental data from single cell analyses (published in EMBO Journal 2006).
Sensitivity Analysis: This option analyses how the up- or down-regulation of key proteins or the kinetics of cytochrome c-induced apoptosome formation or Smac release influence the time required by effector caspases to cleave their substrate. This mode enables the swift identification of potential key regulatory parameters of apoptosis execution within the signalling network.
1D/2D Substrate Cleavage Profiles: In 1D analysis, detailed temporal substrate cleavage profiles can be predicted for a user defined concentration range of one key protein. In 2D profiles, the model calculates the amount of cleaved substrate for concentration ranges of 2 key proteins. Both modes can be used to identify threshold concentrations that separate conditions that either block apoptotic cell death or allow apoptosis to proceed.
These predictions have the potential to considerably enhance experimental designs by identifying kinetic parameters and protein concentrations that significantly influence signal progression for a given cellular system in advance.
Biological Background:
Apoptosis is an evolutionary conserved, genetically defined form of cell death. Apoptotic events are necessary for maintaining tissue homeostasis and removing superfluous or malignant cells. Enhanced or repressed apoptotic cell death contributes to developmental defects, autoimmune diseases, cancer, and neurological disorders (Meier et al. 2000, Yuan and Yankner 2000)
In most cellular systems, apoptosis is activated through the mitochondrial pathway (Fig. 1). The activation of apoptosis-specific cysteine proteases (caspases) is triggered by a process of mitochondrial outer membrane permeabilization (MOMP). The subsequent release of cytochrome c (cyt-c) induces the oligomerisation of apoptotic protease-activating factor 1 (Apaf-1), procaspase-9, dATP/ATP, and cyt-c into the apoptosome complex (Li et al. 1997, Zou et al. 1997). Apoptosome-bound caspase-9 proteolytically activates effector caspases -3 and -7 which are responsible for most of the morphological and biochemical changes characterising this form of active cell death (Li, et al. 1997, Zou, et al. 1997, Janicke et al. 1998, Slee et al. 1999). Caspases-3, -7, and -9 can be inhibited by X-linked-inhibitor-of-apoptosis-protein (XIAP), the most potent representative of the inhibitor-of-apoptosis-protein family (Deveraux and Reed 1999). Furthermore, XIAP enforces the proteasomal degradation of its binding partners by ubiquitination (MacFarlane et al. 2002, Vaux and Silke 2005). Adding complexity, XIAP can be cleaved by active caspase-3 and can also be antagonized by Smac/DIABLO, another protein released from the mitochondria together with cyt-c (Deveraux et al. 1999, Du et al. 2000, Verhagen et al. 2000, Rehm et al. 2003)). The signalling triggered by MOMP therefore induces a complex network of interactions and feed-backs where multiple variables act at the same time.
A full description of the original modelling approach for HeLa cells can be found here.
References:
Deveraux, Q.L. et al. (1999) Cleavage of human inhibitor of apoptosis protein XIAP results in fragments with distinct specificities for caspases, Embo J, 18, 5242-5251.
Deveraux, Q.L. et al. (1999) IAP family proteins–suppressors of apoptosis, Genes Dev, 13, 239-252.
Du, C. et al. (2000) Smac, a mitochondrial protein that promotes cytochrome c-dependent caspase activation by eliminating IAP inhibition, Cell, 102, 33-42.
Goldstein, J.C. et al. (2000) The coordinate release of cytochrome c during apoptosis is rapid, complete and kinetically invariant, Nat Cell Biol, 2, 156-162.
Janicke, R.U. et al. (1998) Caspase-3 is required for DNA fragmentation and morphological changes associated with apoptosis, J Biol Chem, 273, 9357-9360.
Li, P. et al. (1997) Cytochrome c and dATP-dependent formation of Apaf-1/caspase-9 complex initiates an apoptotic protease cascade, Cell, 91, 479-489.
MacFarlane, M. et al. (2002) Proteasome-mediated degradation of Smac during apoptosis: XIAP promotes Smac ubiquitination in vitro, J Biol Chem, 277, 36611-36616.
Meier, P. et al. (2000) Apoptosis in development, Nature, 407, 796-801.
Rehm, M. et al. (2002) Single-cell fluorescence resonance energy transfer analysis demonstrates that caspase activation during apoptosis is a rapid process. Role of caspase-3, J Biol Chem, 277, 24506-24514.
Rehm, M. et al. (2003) Real-time single cell analysis of Smac/DIABLO release during apoptosis, J Cell Biol, 162, 1031-1043.
Slee, E.A. et al. (1999) Ordering the cytochrome c-initiated caspase cascade: hierarchical activation of caspases-2, -3, -6, -7, -8, and -10 in a caspase-9-dependent manner, J Cell Biol, 144, 281-292.
Tyas, L. et al. (2000) Rapid caspase-3 activation during apoptosis revealed using fluorescence-resonance energy transfer, EMBO rep, 1, 266-270.
Vaux, D.L. et al. (2005) IAPs, RINGs and ubiquitylation, Nat Rev Mol Cell Biol, 6, 287-297.
Verhagen, A.M. et al. (2000) Identification of DIABLO, a mammalian protein that promotes apoptosis by binding to and antagonizing IAP proteins, Cell, 102, 43-53.
Yuan, J. et al. (2000) Apoptosis in the nervous system, Nature, 407, 802-809.
Zou, H. et al. (1997) Apaf-1, a human protein homologous to C. elegans CED-4, participates in cytochrome c-dependent activation of caspase-3, Cell, 90, 405-413.
Modelling Background
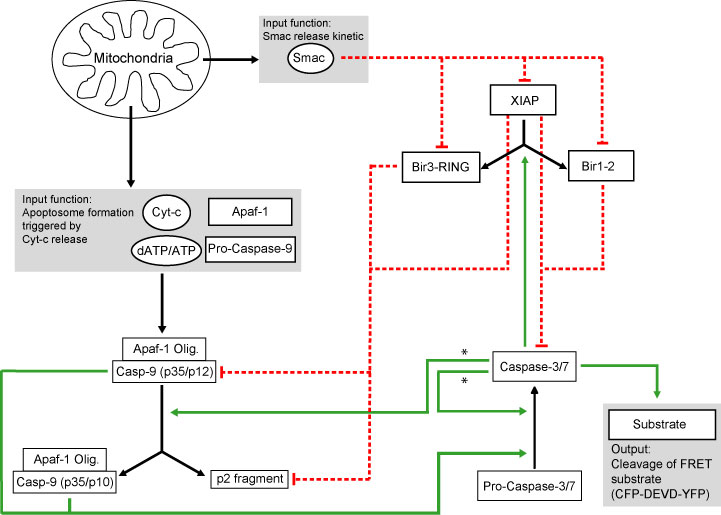
The model is initiated by input functions that remodel cyt-c and Smac release, and apoptosome formation following MOMP. These inputs trigger a network of biochemical reactions that are based on the law of mass action. Moreover, proteasomal degradation and protein production rates were implemented. This network led to a set of 52 ordinary differential equations with 19 reaction partners. As ultimately effector caspase activity orchestrates cell death by cleavage of key cellular substrates, such substrate cleavage was implemented as a model output. Importantly, both MOMP and substrate cleavage kinetics can be measured experimentally in single living cells using established fluorescent probes like tetramethyl-rhodamine-methylester, GFP-fusions with cyt-c or Smac, or effector caspase specific FRET probes (Goldstein et al. 2000, Tyas et al. 2000, Rehm et al. 2002, Rehm, et al. 2003). This enables a direct comparison of the computational predictions to experimental results.
A full description of the original modelling approach for HeLa cells can be found here.
References:
Goldstein, J.C. et al. (2000) The coordinate release of cytochrome c during apoptosis is rapid, complete and kinetically invariant, Nat Cell Biol, 2, 156-162.
Rehm, M. et al. (2002) Single-cell fluorescence resonance energy transfer analysis demonstrates that caspase activation during apoptosis is a rapid process. Role of caspase-3, J Biol Chem, 277, 24506-24514.
Rehm, M. et al. (2003) Real-time single cell analysis of Smac/DIABLO release during apoptosis, J Cell Biol, 162, 1031-1043.
Tyas, L. et al. (2000) Rapid caspase-3 activation during apoptosis revealed using fluorescence-resonance energy transfer, EMBO rep, 1, 266-270.
Technical Implementation
Model equations are solved by MATLAB’s ordinary differential equation solver ODE15 using an adaptive step Runge–Kutta method. The cellular data base is implemented as a flat file maintained by a PERL script.
Database and Submission of quantitative cellular data
Cell type specific quantitative data for the variable parameters used in the model are stored in a database structure. After the selection of a specific cell type in the left frame, values for each parameter can be selected from a drop down menu. Automatically information on the submitting person (name, linked email, affiliation), publication reference (Pubmed ID, if available), and comments on how this value was obtained are displayed. Using the “fill form” button these values can automatically be inputted into the respective calculation forms.
The submission page allows submitting data for new cell types or additional data on cell types already existing in the database. Instead of a full description of all cellular parameters, the system allows the submission of single parameters. Notably, even if a parameter for a specific cell line was already submitted before it is possible to submit an additional value that might differ from the previous entry. Upon refreshing the page all entries will appear in the drop down menus for the respective cell lines. Collecting these entries in the future will allow to display the variability in parameter quantifications arising from different experimental approaches.