We at the CSM are focused on developing tools to deliver personalized risk stratification and treatment response in cancer patients (predominantly for colorectal and glioblastoma). We build network models, integrate multiple high-throughput omic datasets and apply statistical and machine learning algorithms to examine the dysregulation of biological pathways in cancer. We employ these computational tools on datasets from cell lines, animal models and patient samples to systematically interrogate pathways and identify case-specific critical vulnerabilities that could be targeted therapeutically. I collaborate with experimentalists, statisticians, bioinformaticians and clinicians to design experiments to calibrate and test computational predictions, collect and harmonize clinical information and design analysis pipelines.
Modelling tools are developed on the basis of
- Ordinary/Partial Differential equations (ODE/PDE),
- Cellular Automata (CA),
- Decision Trees,
- Methods form control theory, and
- Tools using statistical inference.
Computational models are implemented in programming languages such as MATLAB, MATHEMATICA, C++, or R.
Several tools have been filed for European and US patents. These tools include
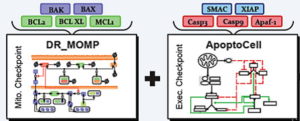
- Dose-Response Medicinal Outcome Model Predictor (DR_MOMP). A tool for evaluating dosage decisions in chemotherapies involving genotoxic stress
- APOPTO-CELL: A tool evaluating apoptosis impairment caused by de-regulation of protein expressions in the mitochondrial pathway subsequent to mitochondrial outer membrane permeabilisation.
- Systems Simulations for the personalisation of Anti-Cancer Treatments (SYS-ACT): Predicting apoptosis susceptibility using a novel approach of knowledge- and data-driven modelling.
- Automated Life-cell Imaging System for Signal-transduction Analyses (ALISSA): An intelligent microscopy system that allows studying signal transduction and that helps to acquire data in a high resolution.
Below are descriptions of the tools and Techniques employed in the CSM
- DR_MOMP
Dose-Response Medicinal Outcome Model Predictor - SYS-ACT
A method for knowledge- and data-driven Systems Simulations for the personalization and optimization of Anti-Cancer Treatments - ALISSA
Automated Life-Cell Imaging System for Signal-transduction Analyses - APOPTO-Cell / PCCP
a clinical workflow and software tool for the Prediction of Cancer response to genotoxic Chemotherapy and Personalised neoadjuvant treatments (PCCP) - TOXI-sim
Analysis of mitochondrial and plasma membrane potentials - miRNAmeConverter
convert mature miRNA names to different miRBase versions
Publications
click here for all the Centre for Systems Medicine publications using these tools