Biological Background
Cells are able to undergo a selective and regulated suicide which is called programmed cell death or apoptosis, which is in sharp contrast to necrosis. Apoptosis has an essential role in the process to form the body shape during embryonic development, but also plays a major role in holding the balance between cell division and cell death.
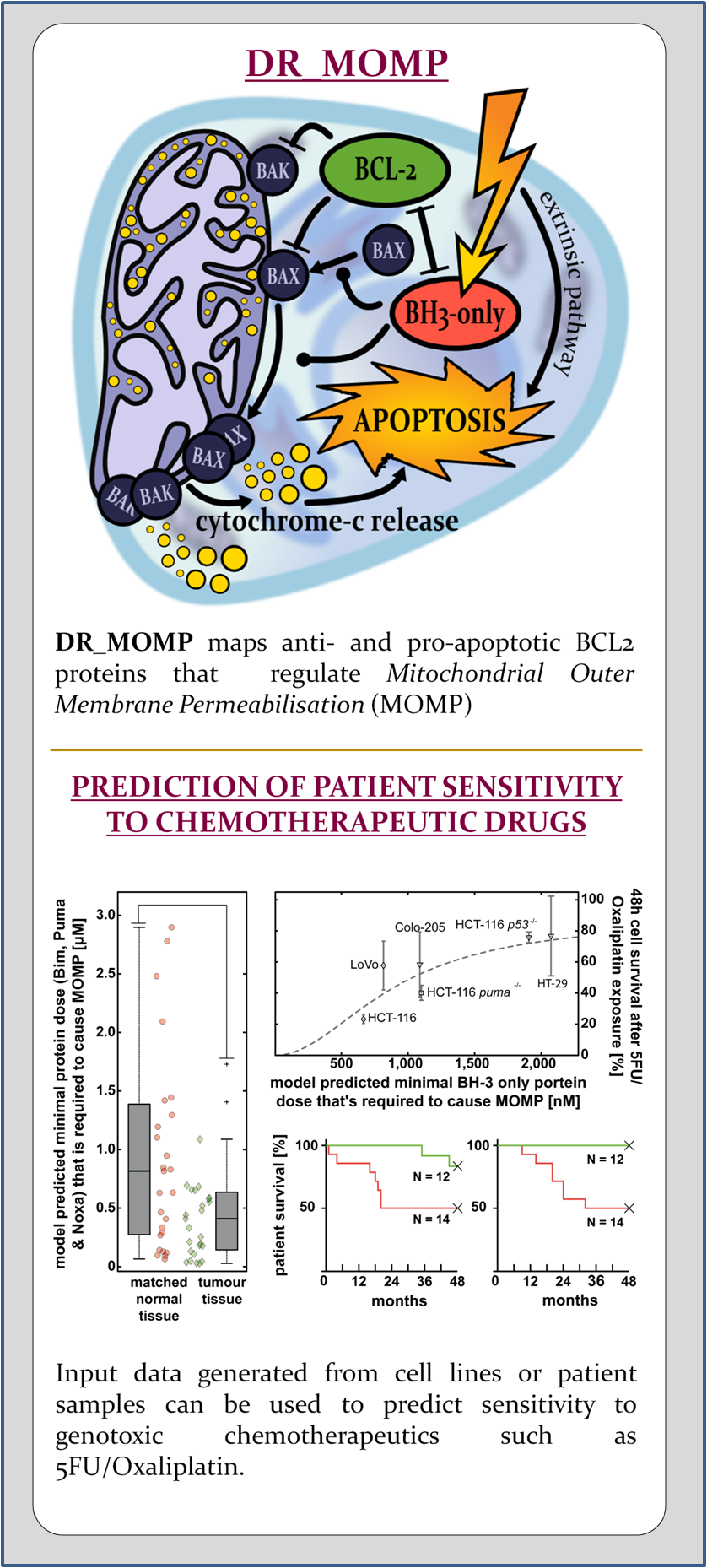
Apoptosis is regulated by various signaling pathways. One pivotal role has the mitochondrial pathway which is regulated by the BCL-2 family of proteins. The BCL-2 family of proteins are grouped into pro- and anti-apoptotic proteins which antagonise each other. Preponderance of pro-apoptotic proteins eventually lead to the mitochondrial outer membrane permeabilisation (MOMP), which is seen as the point of no return in apoptosis. The pro-apoptotic BCL–2 family members BAK and BAX play a direct role in the cause of MOMP and their simultaneous loss leads to impairment of apoptosis. BH3-only proteins are pro-apoptotic and are up-regulated or activated by various stimuli. Typical intrinsic stimuli that are capable to induce the activation of BH3-only proteins are DNA damage, anoikis or hypoxia. BH3–nly proteins inhibit anti-apoptotic BCL-2 family proteins which in turn inhibit the pro-apoptotic effector proteins BAK and BAX. Free and active BAK and BAX homo-oligomerise and subsequently induce MOMP, which allows the release of mitochondrial proteins such as diablo IAP-binding mitochondrial protein (DIABLO, also called SMAC) or cytochrome-c (Cyto-c) to the cytosol. There, Cyto–c forms with the apoptotic peptidase activating factor~1 (APAF1) the apoptosome. The apoptosome is a platform that facilitates the cleavage of procaspase-9 to active caspase-9 (see APOPTO-CELL ).
DR_MOMP is a computational model representing the family of BCL-2 proteins and their protein interactions. The model implements the direct activation model of the effectors proteins BAK and BAX by the activator proteins BIM, tBID and PUMA. Protein interactions were modeled based on the law of mass action.
We created DR_MOMP to evaluate the pro- and anti-apoptotic BCL2 family proteins, and their regulation of mitochondrial outer membrane permeabilisation (MOMP). Using DR_MOMP and absolute protein levels of the proteins BAK, BAX, BCL2, BCL(X)L and MCL1, we are able to calculate a stress dose required to induce MOMP and eventually cell death. The stress dose is defined by BH3-only proteins and its composition can be adapted to individual needs. TNF-related apoptosis-inducing ligand (TRAIL) signaling can be simulated using tBID as component. Genotoxic sterss (for example caused by UV radiation, 5-fluorouracil, cisplatin, capecitabine, oxaliplatin, paclitaxel, etcetera) can be modeled using BBC3 (PUMA), BIM and NOXA (PMAIP1). Removing BBC3 allows to account for tp53 mutations.
Cells’ calculated stress doses positively correlate with % cell survival in vivo experiments and allows us to assess whether or not cells are sensitive to apoptotic stress triggered by chemotherapy. In patients, primary tumour tissue with a high stress dose is associated with unfavourable clinical outcome, due to cancer cells resistance to undergo apoptosis. In contrast, tumour tissue with a low stress dose which is associated with favourable clinical outcome, due to high cancer cell death rates.
DR_MOMP’s predictive efficiency was thoroughly tested with colorectal and triple negative breast cancer cell lines in vivo, and colon- and rectal cancer in retrospective studies with adjuvant and neoadjuvant chemotherpy settings.
Link to https://www.systemsmedicineireland.ie/tools/apopto-cell/
- Lucantoni et al. Cell Death Dis. 2018 Jan 19;9(2):42. doi: 10.1038/s41419-017-0039-y. PMID: 29352235
- Lindner et al. 2017 Dec;66(12):2141-2148. doi: 10.1136/gutjnl-2016-312287. PMID: 27663504
- Flanagan et al. J Mol Med (Berl). 2015 Mar;93(3):315-26. doi: 10.1007/s00109-014-1221-7. PMID: 25388617
- Lindner et al. Cancer Res. 2013 Jan 15;73(2):519-28. doi: 10.1158/0008-5472.CAN-12-2269. PMID: 23329644
References
- Shi et al. Mol Cell. 2002 Mar;9(3):459-70. PMID: 11931755.
- Benn et al. Nat Rev Neurosci. 2004 Sep;5(9):686-700. PMID: 15322527)
- Willis et al. Curr Opin Cell Biol. 2005 Dec;17(6):617-25. PMID: 16243507
- Singh et al. Nat Rev Mol Cell Biol. 2019 Mar;20(3):175-193. doi: 10.1038/s41580-018-0089-8.. PMID: 30655609
https://www.ncbi.nlm.nih.gov/pubmed/29352235
https://www.ncbi.nlm.nih.gov/pubmed/27663504
https://www.ncbi.nlm.nih.gov/pubmed/25388617
https://www.ncbi.nlm.nih.gov/pubmed/23329644
https://www.ncbi.nlm.nih.gov/pubmed/11931755
https://www.ncbi.nlm.nih.gov/pubmed/15322527
https://www.ncbi.nlm.nih.gov/pubmed/16243507
https://www.ncbi.nlm.nih.gov/pubmed/30655609